Transgressive segregation is the phenomenon in which segregating hybrids exhibit phenotypes that are extreme or novel relative to the parental lines, and typically results from epistatic interactions between the genes assembled in novel combinations. Cotton belongs to allotetraploid and homoeologous chromosomes make recombination more complexed. Professor Wang from Pest Control Group has been collaborating with Professor Philip A. Roberts from University of California, Riverside for many years. Their previous studies indicated that the RKN resistance trait in tetraploid cotton is not like other single gene or two-gene trait which can segregate in a traditional Mendelian law and continually confirmed that transgressive resistance to root-knot nematode in cotton is very common in progenies derived from combinations of different genomes (Wang et al. 2006, 2008, 2012, 2015, 2017). Importantly, they found that transgressive factors from susceptible parental lines contributed to super resistance and these were mapped to the same region as a known rkn1 resistance gene on Chr11 containing resistance rich-gene clusters, yet its homoeologous Chr21 has no contribution to nematode resistance but with similar gene order.
To reveal the transgressive mechanism at the genomic level, this study represents that the first time sequence comparisons between Chr11 and Chr21 were made in the nematode resistant line since all published cotton genomes are susceptible to root-knot nematode. The result of this study indicated that less mapped sequence with nematode susceptible TM1 reference genome was identified in the rkn1 region on Chr11 than that on Chr21 and the relocation of the markers within Chr11 or Chr21 and between Chr11 and Chr21 was observed in the genetic linkage map and TM-1 physical map (Figure 1) which might explain the differences in resistance between the pair of chromosomes. More multiple homologous copies of R proteins (Figure 2) and adjacent transposable elements are present within Chr11 than within Chr21, indicating transposable elements might be involved in nematode resistance or transgressive resistance in the rkn1 region. The identified unique insertion/deletion in NB-ARC domain, different copies of LRR domain in the resistant cultivar (Figure 3) might be also major factors contributing to complex recombination and transgressive resistance which will be a prime target for following to study gene function. We believe this system and our findings provide a model to study transgressive resistance in plants. Also, the further functional characterization of these R protein domains will shed light on resistance evolution and be helpful for developing new sources of resistant crops.
This study was published in the international journal “Frontiers in Plant Science”. The first author is Dr. Congli Wang and the corresponding author is Dr. Philip A. Roberts. The research was partially supported by National Natural Science Foundation of China.
Paper: Wang C, Ulloa M, Nichols RL and Roberts PA (2020) Sequence Composition of Bacterial Chromosome Clones in a Transgressive Root-Knot Nematode Resistance Chromosome Region in Tetraploid Cotton. Front. Plant Sci. 11:574486. https://doi.org/10.3389/fpls.2020.574486
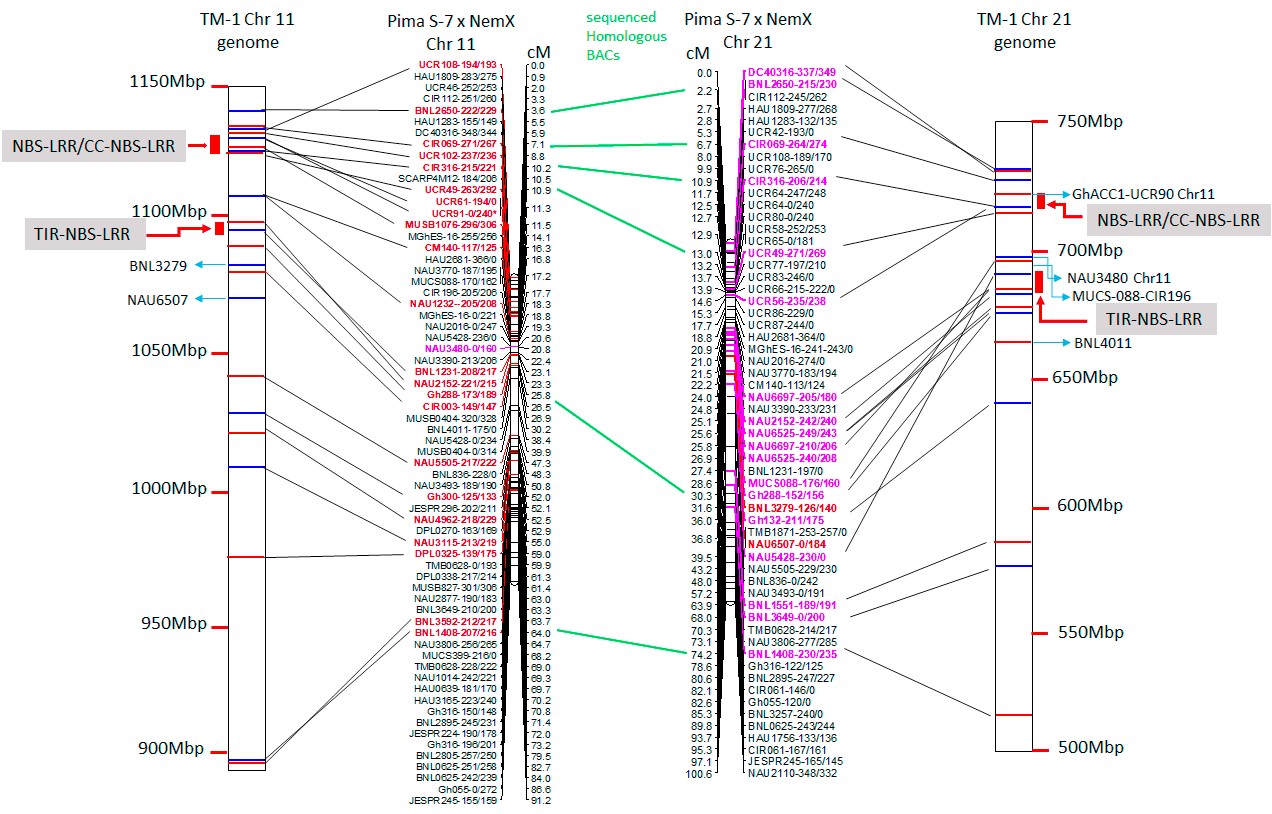
Figure 1. Alignment of TM-1 reference genome (https://www.cottongen.org/analysis/189) with linkage maps of Chr11 and its homoeologous Chr21 using an interspecific [Pima S-7 (Gossypium barbadense) x Upland Acala NemX (G. hirsutum)] RIL population (Wang et al., 2017). SSR markers in red are mapped to TM-1 Chr11 genome and those in pink are mapped to TM-1 Chr21 genome.
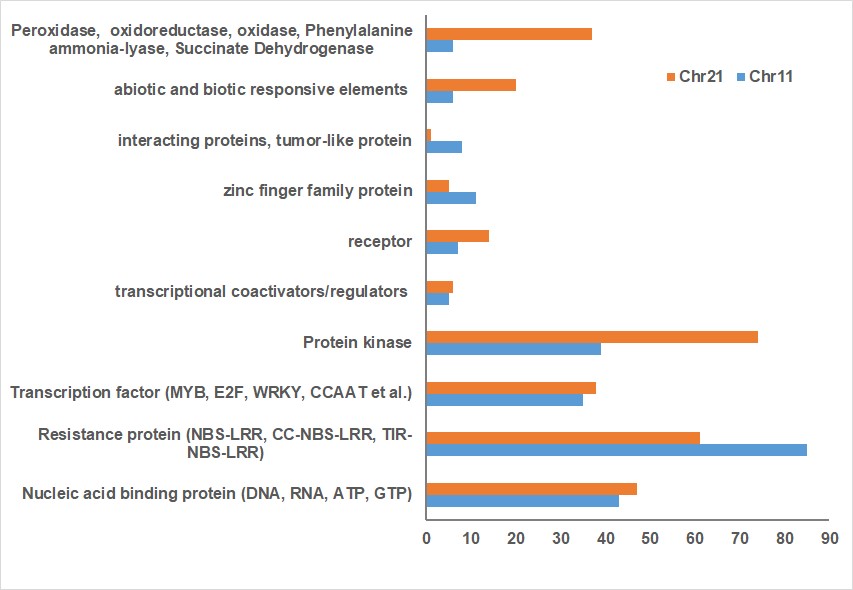
Figure 2: Comparison of number of stress response elements on Chr11 BACs with those on Chr21 BACs based on Gossypium hirsutum Unigene NCBI database.
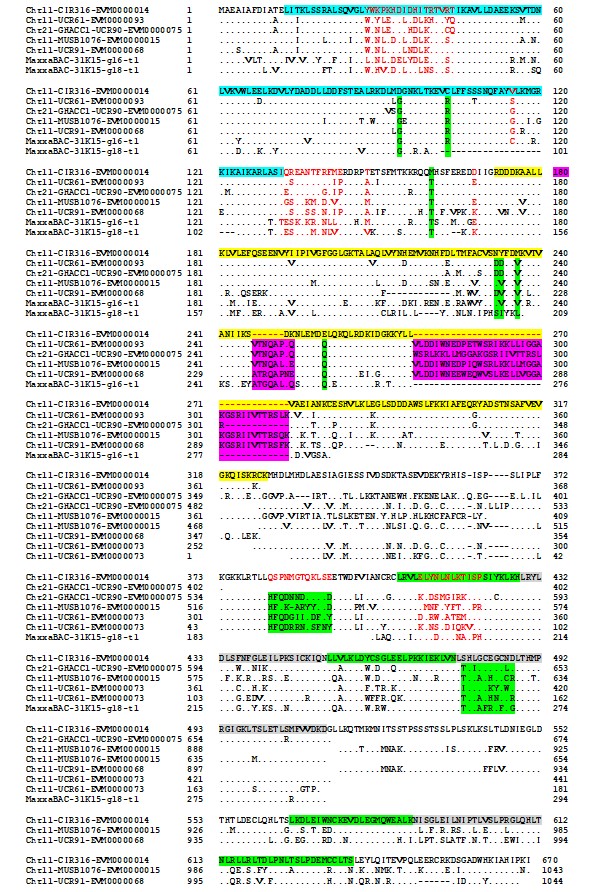
Figure 3: Alignment of disease resistance protein (CC)-NBS-LRR among BACs on Acala N901 Chr11-21 BACs and Acala Maxxa BAC 31K15 (Wang et al., 2015) in the rkn1 region. The sequence in light blue in the first line represents CC domain, with NB-ARC domain in yellow and LRR domains in green and gray.

