Plastic films is one of the most sources of terrestrial microplastics (MPs), creating distinct microbial niches from soils, termed as plastisphere. However, there are considerable gaps in our understanding on the mechanisms of MPs on soil N transformation and bioavailability by changing microbial functional profiles.In a study recently published in Journal of Hazardous Materials, researchers from WANG Guanghua’s group at the Northeast Institute of Geography and Agroecology, Key Laboratory of Mollisols Agroecology, Chinese Academy of Sciences (CAS) revealed that addition of conventional and biodegradable MPs significantly shifted the microbial functional and genomic information encoding nitrogen (N) cycling within the plastispheres and soils.
By metagenomics and genome binning methods, this study reported that biodegradable MPs harbored rougher surfaces and induced stronger alterations in microbial functional and taxonomic profiles in the soil and plastisphere than conventional ones. In comparison to their respective soils, the plastispheres of conventional and biodegradable MPs stimulated the processes of nitrogen fixation, nitrogen degradation and assimilatory nitrate reduction (ANRA) and reduced the gene abundances encoding nitrification and denitrification, in which biodegradable MPs induced stronger influences.
Biodegradable plastisphere harbored higher relative abundances of Proteobacteria and some genera affiliated with this phylum, such as Sphingomonas, Ramlibacter Variovorax and Caenimonas, carrying N cycle-related functional genes. Differently, conventional plastisphere enriched Actinobacteria together with Nocardia and Amycolatopsis affiliated with it.
Moreover, Ramlibacter mainly drove the differences in N cycling processes between the soils containing two types of MPs and was further enriched in the BMP plastisphere. Three genomes of Ramlibacter contained diverse functional genes for N cycling were reconstructed. The novel Ramlibacter strains had the potential capacities to generate ammonium through multiple metabolic routes, supporting their biosynthesis and finally resulted in the accumulation of soil NH4+-N(Figure 1).
This study provides new insights that the genetic mechanisms of soil N bioavailability in the presence of biodegradable MPs, which have important implications for maintaining sustainable agriculture and controlling microplastic risk.
The study was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDA28020201) and Heilongjiang Provincial Natural Science Foundation of China (ZD2022D001).
The finding of this study was recently published online in Journal of Hazardous Materials. https://www.sciencedirect.com/science/article/pii/S0304389423003795.
Key words: Microplastics, Plastisphere, Metagenomics, Nitrogen cycling, Metagenome-assembled genome
The full information of the article:
Hu X, Gu H, Sun X, Wang Y, Liu J, Yu Z, Li Y, Jin J, Wang G*. Distinct influence of conventional and biodegradable microplastics on microbe-driving nitrogen cycling processes in soils and plastispheres as evaluated by metagenomic analysis. Journal of Hazardous Materials. 2023, 131097.
doi: 10.1016/j.jhazmat.2023.131097.
Contact:
Prof. WANG Guanghua, Ph.D Principal Investigator
Northeast Institute of Geography and Agroecology, Key Laboratory of Mollisols Agroecology, Chinese Academy of Sciences, Harbin 150081, China
E-mail: wanggh@iga.ac.cn
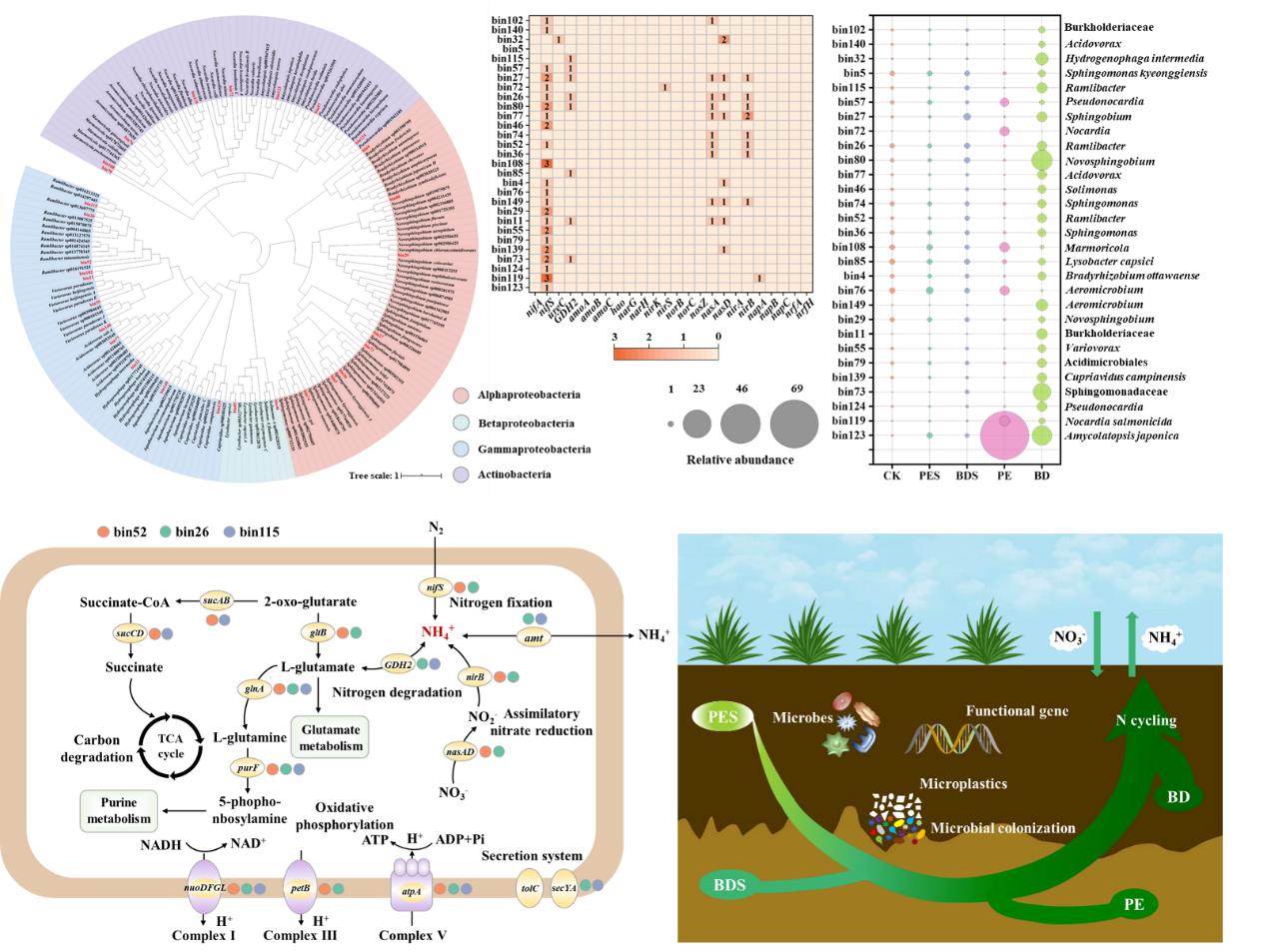
Figure 1. The influences of conventional and biodegradable microplastics on microbial nitrogen functional profiles within soils and plastispheres. (Image by HU Xiaojing)

