Recently, a research team led by Prof. WANG Guanghua from the Northeast Institute of Geography and Agroecology, Key Laboratory of Mollisols Agroecology, Chinese Academy of Sciences (CAS) revealed that microbial adaptive evolution and regulatory strategies in response to soil carbon (C) heterogeneity in paddy soils at spatial scales.
This work was published in Geoderma on 17 March 2025.
Paddy soils, as important carbon sinks, are 39%-127% more efficient in sequestration of SOC compared to upland soils. However, the adaptive evolution and regulatory strategies of microorganisms in response to soil carbon heterogeneity in paddy soils at spatial scales remain poorly understood.
According to the researchers, significant distance-decay relationships (DDRs) were observed at both microbial C functional and genomic taxonomic levels. Microbial C cycling profiles were clustered into two groups. HCS (including sites R1-R10) represented soils with high total carbon (TC) at relatively high latitudes, whereas LCS (including sites R11-R30) had low soil C content distributed at low latitudes. Compared with HCS, LCS presented higher abundances of C cycling pathways involving aerobic respiration, C fixation, and methanogenesis pathways, as well as higher levels of carbohydrate esterase (CE) and glycosyl transferase (GT) classes.
211 metagenome-assembled genomes (MAGs) with diverse C metabolic functions were constructed. Among these, high-quality MAG292, assigned to the Nanopelagicales order, had a significantly positive correlation with TC and was more abundant in HCS. Contrarily, MAG153, assigned to the Chitinophagales order, exhibited an opposite trend.
Additionally, 133 novel vMAGs were retrieved, and the abundances of phage11, phage16, phage26, and phage120 were higher in LCS than in HCS, containing chiA and GH19 that involved in chitin degradation. HCS had a relatively high abundance of phage89, containing slt and GH23 genes that regulate peptidoglycan lysis.
These results indicated that soil viruses potentially lyse bacteria by encoding peptidoglycan lyase, releasing nutrients, and increasing the amount of dead microbial debris that facilitates soil C accumulation at relatively high latitudes. In contrast, at low latitudes, the phages together with microbes may indirectly decrease the soil TC by potentially expressing auxiliary metabolic genes (AMGs) involved in chitin degradation.
It highlights the divergent microbial adaptive evolution and soil C regulation strategies response to soil C heterogeneity in paddy soils of Chinese Mollisols.
This study was supported by the National Key Research and Development Program of China (2022YFD1500201) and the Youth Innovation Promotion Association of Chinese Academy of Sciences (2023237).
The finding of this study was recently published online in Geoderma.
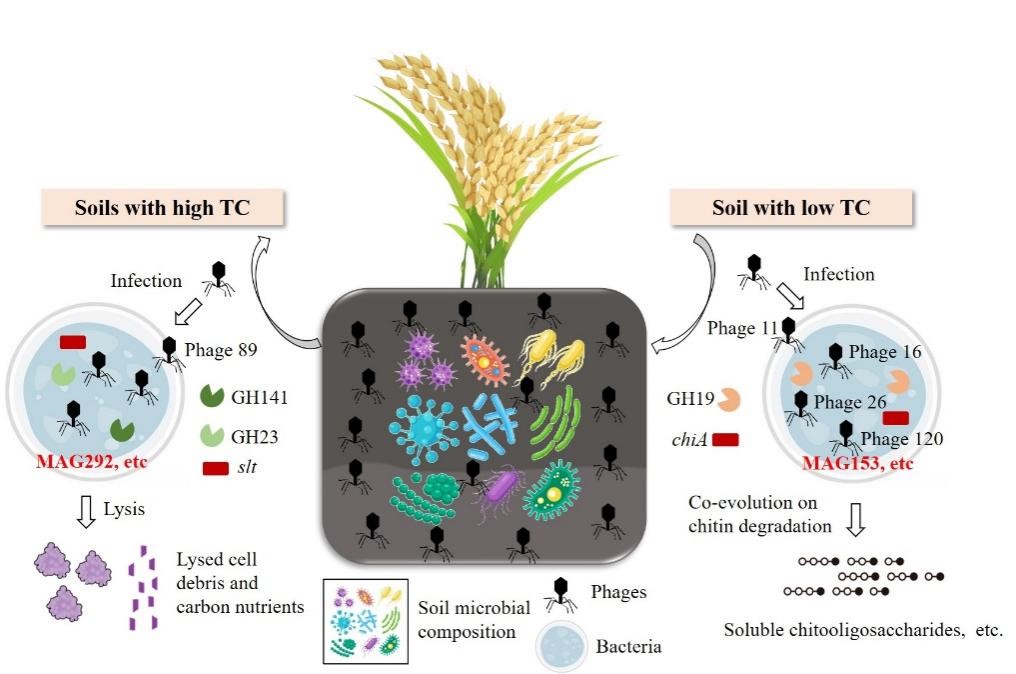
Figure 1. Schematic showing the adaptive and regulatory mechanisms of microbes and potentially associated viruses with soil carbon heterogeneity in paddy soils. (Image by HU Xiaojing)
https://doi.org/10.1016/j.geoderma.2025.117265.
Key words: Microbial biogeography, Carbon cycling pathways, Carbohydrate-active enzymes, Metagenome-assembled genomes, Viral genome
The full information of the article:
Xiaojing Hu, Haidong Gu, Mikhail Semenov, Yongbin Wang, Jinyuan Zhang, Zhenhua Yu, Yansheng Li, Junjie Liu, Jian Jin, Xiaobing Liu, Guanghua Wang. Biogeographic patterns and adaptive strategies of microbial carbon metabolic profiles in paddy soils in the Chinese Mollisol region. Geoderma. 2025, 117265.
Contact:
Prof. WANG Guanghua, Ph.D Principal Investigator
Northeast Institute of Geography and Agroecology, Key Laboratory of Mollisols Agroecology, Chinese Academy of Sciences, Harbin 150081, China
E-mail: wanggh@iga.ac.cn

